Ezerski
Scientific
Consultations
Ezerski Scientific offers a wide range of consultation services for both small and large biotech companies. We provide predictive analysis for small molecule & oligonucleotide drug development, as well as molecular dynamics modeling for intrinsically disordered proteins/peptides (IDPs), protein-protein interactions, protein folding, kinetics, and more. In addition to the currently available services listed below, we offer customized solutions to meet your business needs.
oligonucleotide efficiency predictions
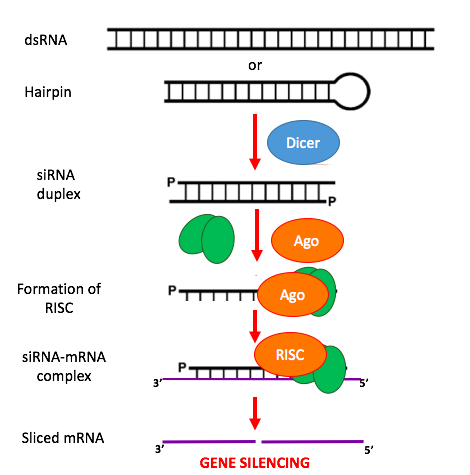
siRNA/saRNA based pharmaceuticals have gained significant attention in recent years due to their vast application in difficult to treat disease indications, genetic disorders, and cancers. Our computational prediction platform can identify the top performing candidates for a transcript of interest.
Biomarker identification
Locating and identifying specific biomarkers are paramount to discovery of diseases and genetic conditions. As a result, the use of biomarkers has become an essential tool in drug development. Due to the extreme size of genetic datasets, efficient computational tools are necessary to handle this task. Ezerski Scientific will let you skip the costly development phase and find patterns and potential biomarkers usinging our advanced algorithms, so your organization can focus on generating results.
Molecular dynamics simulaton
Modeling receptor-ligand, protein-protein, or equilibrium ensemble structures is an extremely difficult task that requires the use of MD simulation technology. This computational method enables the observation of molecular interactions that are unobtainable through experimental means. The results of MD simulations are able to generate realistic kinetic predictions.
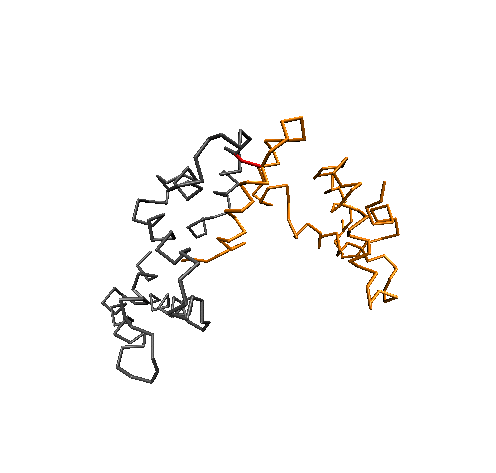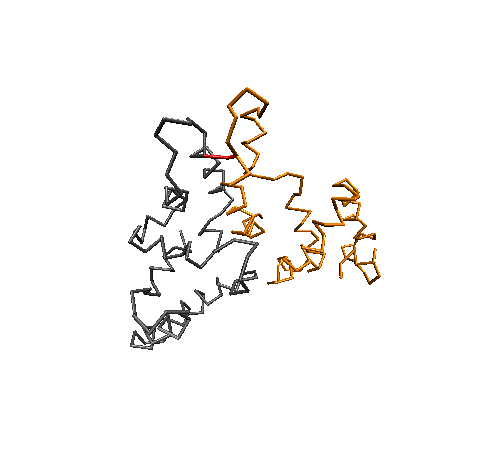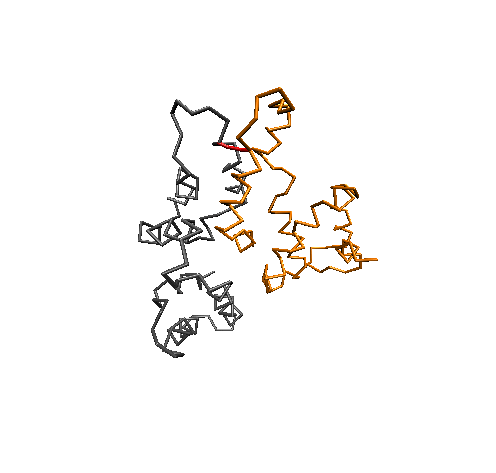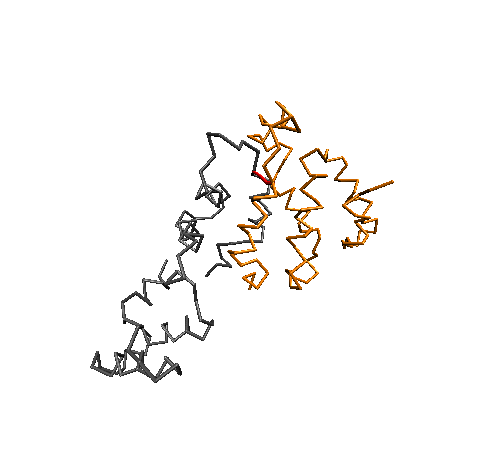
Conformational Clustering
MD simulations offer an atomistic level of resolution, which is essential for functional analysis of IDPs, proteins, receptors, and more. In addition to standard methods, we utilize the Combinatorial Average Transient Structure (CATS) algorithm for determining structural ensembles of IDPs. The clusters generated by CATS require no a priori information about the expected structure, which is advantageous for determining dominant structures in MD trajectories when experimentally determined structures are unavailable.
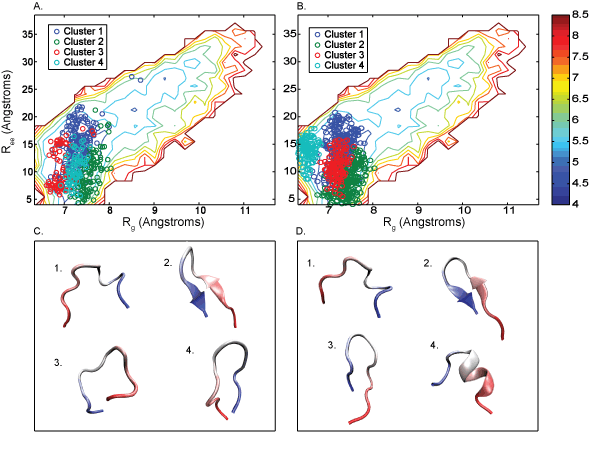
Markov State Modeling
The kinetics of equilibrium protein binding can be modeled using Markov State Model (MSM). The MSM is particularly useful in illustrating distinct binding pathways that are responsible for observed experimental protein kinetics. This offers a distinct advantage over modeling kinetics via free energy in complex protein interactions.
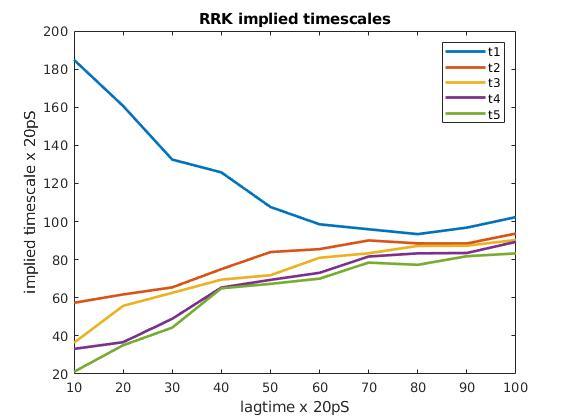
Custom SOlutions
Pushing the boundaries of science sometimes requires a novel approach. Ezerski Scientific can help. We apply our expertise in AI/ML, molecular dynamics simulation, data science, statistics, bioinformatics and algorithm development to satisfy the needs of our clients. Our customized solutions are guaranteed for the lifetime of your project and comes with unlimited technical support.